GlimmerHMM User Manual
I. OverviewGlimmerHMM is a new gene
finder based on a Generalized Hidden Markov Model (GHMM). Although the
gene finder conforms to the overall mathematical framework of a GHMM,
additionally it incorporates splice site models adapted from the GeneSplicer
program.
The variable-length feature states (e.g., exons, introns, intergenic regions) are
implemented using Nth-order interpolated Markov
models (IMM) as described in Delcher et al.
1999 for N=8. Currently, GlimmerHMM's
GHMM structure includes introns of each phase, intergenic regions, and
four types of exons (initial, internal, final, and single), as depicted
in the figure below. 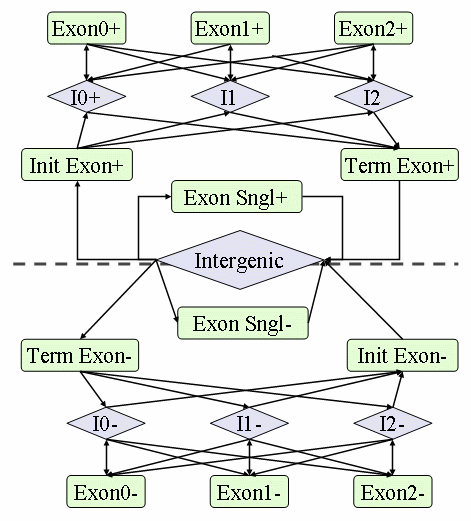 There are
a number of assumptions used by GlimmerHMM when predicting genes in the
DNA sequence. (1) the coding region of every gene begins with a
start codon ATG (but partial genes can be predicted), (2) a gene has no in-frame stop codons except the very last codon, (3) each exon is in a consistent reading frame with the previous exon. These constraints significantly enhance the efficiency of computing the optimal gene models, by restricting the search space of the GHMM algorithm. On the other hand, genuine frame shifts cannot be detected by the system. III.Training an organism specific GlimmerHMM
III.1. Need of a specific organism trainingOur earlier implementation of a gene finder - GlimmerM - trained for rice produced clearly superior gene models than GlimmerM trained for Arabidopsis (Pertea and Salzberg 2002). This is also valid for GlimmerHMM. Although the current trainings of GlimmerHMM should work well on closely related organisms, the user should use our training procedure to re-train the system for an other eukaryotic organism in order to improve the accuracy of the gene prediction. For all species that GlimmerHMM is already trained, we expect the performance to improve even further by re-training the system as the amount of DNA sequences from these species continues to grow. III.2 Data collectionFirst of all it should be noted that a thorough collection of a good training data set is a step that should precede the training of any gene finder. This step is very important since the quality of the data presented for training is directly proportional with the accuracy of the resulted gene finder. As any species-specific gene finder, GlimmerHMM needs a training data set containing as many as possible complete coding sequences from the organism genome for which the gene prediction is intended. By surveying the public databases, one should be able to find all previously known genes of the target organism that are backed by laboratory evidence. These genes would form a sound training data set if their number would be large enough. Unfortunately, this is rarely the case, so one should use other methods in order to construct a reliable data set. From our experience, most often the training data set was formed by searching long ORFs (with more than 500 bases) against non redundant protein sequence databases by using programs like BLAST in order to map known proteins into the genome. III.3 Training GlimmerHMMTo train GlimmerM you should run trainGlimmerM with the following command: trainGlimmerHMM <mfasta_file> <exon_file> [optional_parameters] <mfasta_file>
and <exon_file> are
the
multi-FASTA file and the file containing the exon coordinates of the
known genes, respectively. <mfasta_file>
is a multifasta file containing the sequences for training with the
usual format: If not enough data is available for training the
splice sites the training procedure will be unsuccessful and exit with
a warning message. In this case the user should collect more known
genes with introns and then try the training procedure again. If not
enough data is available for training the translational start and stop
sites of
the genes, the user will need to collect more genes. A minimum number
of 50 genes with standard start codons (ATG) and stop codons
(TAA/TAG/TGA) is assumed by default. The optional parameters that can be given to the
training program are:
III.4 Changing the parameters obtained during training
Choosing adequate thresholds that determine the false negative-false positive ratio for specific site detection may be a challenging problem given the small number of true sites and the overwhelming number of false positives, and requires a suite of decisions that maximizes the accuracy of the recognition task without a big loss in the sensitivity. In the training procedure of GlimmerHMM the threshold for calling a sequence a real splice site is chosen by examining the trade-off in the false positive rate. The system creates a sorted list of thresholds, adjusting the scoring function so that it will miss 1,2,3, etc. true sites. The default threshold is chosen to be the score at which the false positive rate drops by less than 1%. To allow a greater flexibility in setting the signals' thresholds, our training procedure allows the user to consult the false positive and false negative rates specific for the training data and set his own threshold. After running trainGlimmerHMM, a log file can be
consulted to find the default values set for some of the parameters of
GlimmerHMM. The file config.file
in the training directory specifies for each isochore which
configuration file to use. For the default case, when no isochores are
considered, there will be only one line with the foillowing info:
IV. Running GlimmerHMMThe program GlimmerHMM takes two inputs: a DNA
sequence file in FASTA format and a directory containing the training
files for the program. If not specified, the training directory is by
default the current working directory. VI. Obtaining GlimmerHMMGlimmerHMM is available free of charge under the
open-source Artistic License.
To download GlimmerHMM please click here. ReferencesDelcher, A.L., Harmon, D., Kasif, S., White,O. and Salzberg, S.L. Improved microbial gene identification with GLIMMER Nucleic Acids Research, 27:23 (1999), 4636-4641. Majoros, W.H., Pertea, M.,
and Salzberg, S.L.
TigrScan
and GlimmerHMM: two open-source ab initio eukaryotic gene-finders Bioinformatics 20 2878-2879. Majoros,W.M. and Salzberg, S.L. (2004) An
empirical analysis of training protocols for probabilistic gene
finders. BMC Bioinformatics 5:206. Pertea, M., X. Lin, et al. (2001). "GeneSplicer: a new computational method for splice site prediction." Nucleic Acids Res 29(5): 1185-90. Pertea, M. and S. L. Salzberg (2002). "Computational gene finding in plants." Plant Molecular Biology 48(1-2): 39-48. Pertea, M., S. L. Salzberg, et al. (2000). "Finding genes in Plasmodium falciparum." Nature 404(6773): 34; discussion 34-5. Salzberg, S. L., M. Pertea, et al. (1999). "Interpolated Markov models for eukaryotic gene finding." Genomics 59(1): 24-31. Yuan, Q., J. Quackenbush, et al. (2001). "Rice bioinformatics. analysis of rice sequence data and leveraging the data to other plant species." Plant Physiol 125(3): 1166-74. |



