RefSeq annotation of the CHM13 genome
This annotation was created by using the Liftoff program [1]; (v1.6.3, with options -copies -sc 0.95 -polish -exclude_partial -chroms) to map across all human genes in RefSeq [2] annotation release 110 from the GRCh38.p14 genome to the CHM13v2.0 genome, a complete, gap-free human genome published by the Telomere-to-Telomere (T2T) consortium [3].
Of the 58,700 genes (19,871 protein coding) and 179,372 transcripts (130,361 protein coding) annotated on the main chromosomes and unplaced contigs in GRCh38, we successfully lifted over 58,272 genes (19,767 protein coding) and 178,688 transcripts (130,426 protein coding). We also used the "-copies" option in Liftoff to identify additional gene copies in CHM13, using a minimum sequence identity threshold of 95% at the DNA level. With this threshold, we found 2,393 additional gene copies (239 protein coding). Finally, we added the ribosomal DNA (rDNA) annotations on the acrocentric chromosomes from the CHM13v2.0 annotation, which were created using the Comparative Annotation Toolkit (CAT) [4]; and Liftoff. These steps resulted in a total gene count of 61,322 (20,006 protein coding) and a total transcript count of 181,715 (130,426 protein coding). The table below shows the number of genes of each biotype in GRCh38 and CHM13.
| Gene biotype | Number of genes in GRCh38 | Number of genes mapped onto CHM13 |
|---|---|---|
| protein coding | 19871 | 20006 |
| lncRNA | 17793 | 18389 |
| pseudogene | 15357 | 16030 |
| miRNA | 1914 | 2047 |
| transcribed pseudogene | 1221 | 1262 |
| snoRNA | 1195 | 1188 |
| tRNA | 431 | 522 |
| V segment | 239 | 245 |
| V segment pseudogene | 189 | 209 |
| snRNA | 153 | 192 |
| J segment | 98 | 79 |
| ncRNA | 51 | 49 |
| rRNA | 38 | 325 |
| misc RNA | 36 | 37 |
| D segment | 32 | 0 |
| C region | 21 | 23 |
| antisense RNA | 19 | 19 |
| other | 14 | 13 |
| J segment pseudogene | 7 | 6 |
| C region pseudogene | 5 | 5 |
| Y RNA | 4 | 7 |
| vault RNA | 4 | 4 |
| scRNA | 4 | 4 |
| telomerase RNA | 1 | 1 |
| RNase P RNA | 1 | 1 |
| RNase MRP RNA | 1 | 1 |
| ncRNA pseudogene | 1 | 1 |
| Total | 58700 | 60655 |
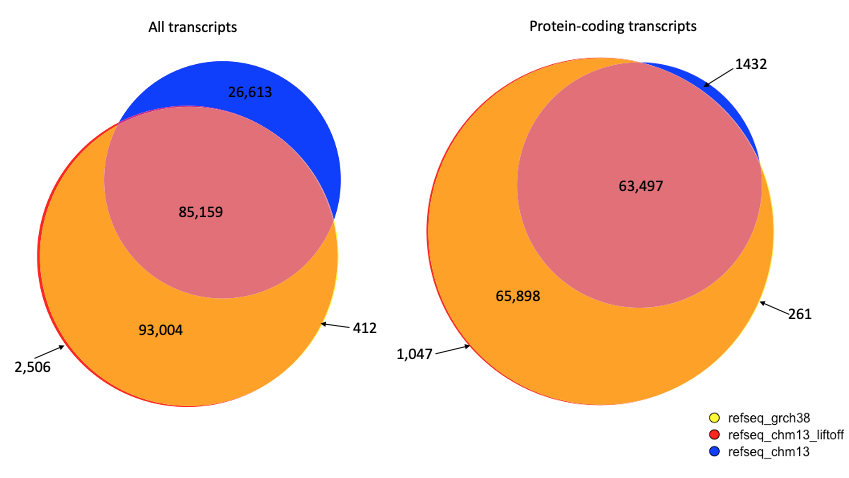
A comparison of the genes using gene IDs to match annotations is shown in the figure below. Genes were compared between RefSeq v110 on GRCh38, the Liftoff version of RefSeq on CHM13, and the NCBI version of RefSeq on CHM13. The NCBI version, shown in blue, was produced using the Gnomon pipeline, which only mapped approximately half of the protein coding genes.
1. Shumate, A. & Salzberg, S. L. Liftoff: accurate mapping of gene annotations. Bioinformatics 37, 1639–1643 (2021).
2. O’Leary, N. A. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44, D733–D745 (2016).
3. Nurk, S. et al. The complete sequence of a human genome. Science (1979) 376, 44–53 (2022).
4. Fiddes, I. T. et al. Comparative Annotation Toolkit (CAT)-simultaneous clade and personal genome annotation. Genome Research 28, 1029–1038 (2018).




